Projects
We develop tools and resources that support our mission to understand how the microbiome and other omic pathways—such as genetics, metabolomics, and epigenetics—influence the response to clinical interventions like diet and exercise. Our projects often emerge from collaborations with clinical studies and large cohort datasets, allowing us to explore diverse populations and generate insights into cardiometabolic health.
Our lab is especially focused on building reproducible, open-source software and statistical workflows that enable the analysis of complex biological data and help advance precision nutrition and personalized health strategies. Below are some of our featured tools and datasets.
Featured
Obesity is a global epidemic that substantially increases risk for numerous chronic diseases. Unfortunately, weight loss (and weight loss maintenance) is very challenging for many people. Some of the reasons for this relate to motivation and the time and cost of maintaining a healthy lifestyle. However, some of the reasons are also physiological in nature and rooted in individual factors, like genetics or the composition of the gut microbiome. We apply advanced computational methods to molecular data from participants of weight loss interventions to understand the effects of these complex physiological factors.
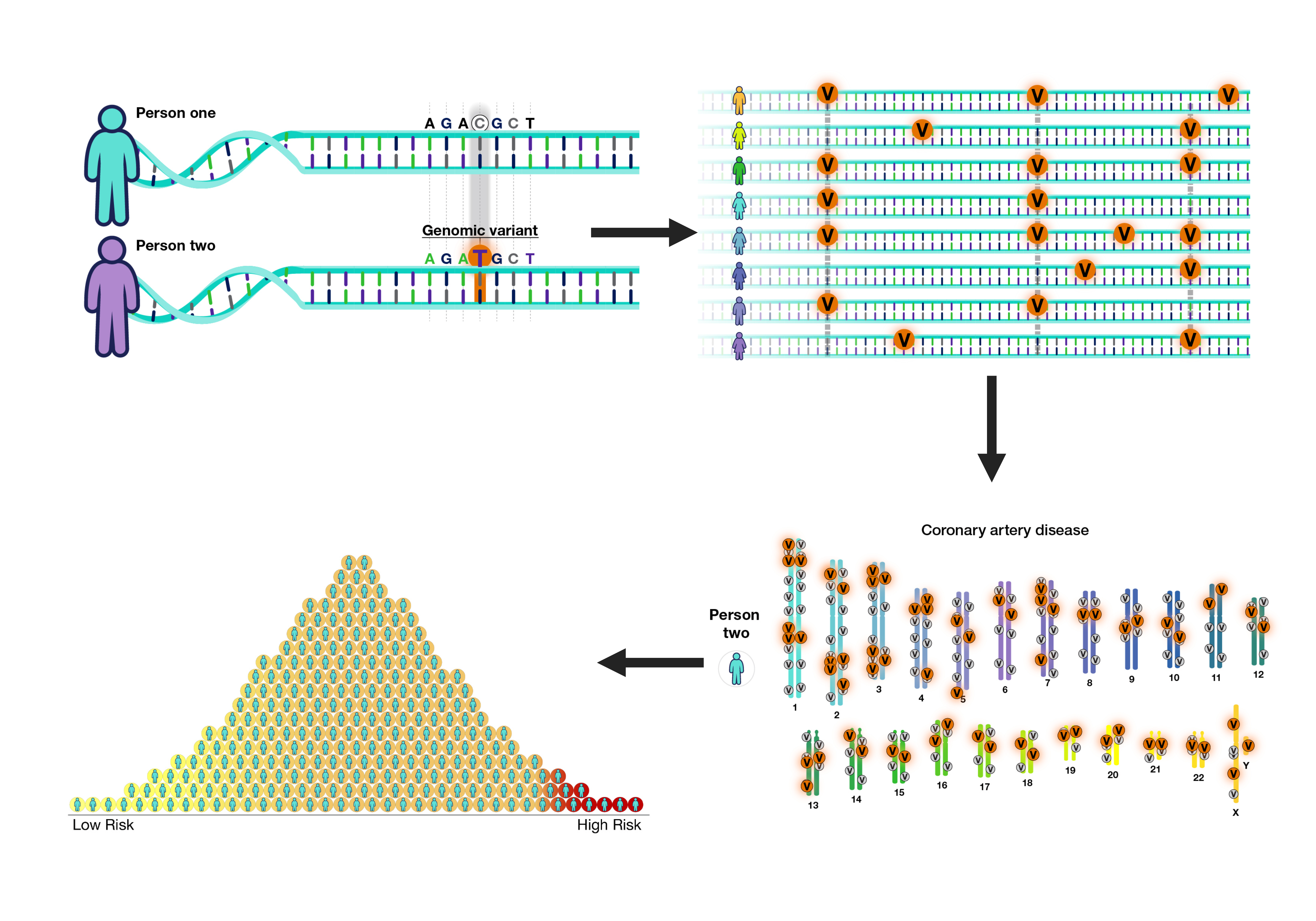
Polygenic risk scores have become a clinically valuable tool to summarize genetic risk for a disease across the genome and identify individuals who are at high or low risk for disease. These are particularly relevant for polygenic diseases, such as obesity or diabetes, where risk variants across numerous genes contribute cumulatively towards disease risk (versus monogenic diseases like cystic fibrosis with risk stemming from one specific gene). Our team applies similar methods to omics data (e.g., data from the microbiome, proteomics, metabolomics, transcriptomics) in relation to various health conditions or disease states, such as inflammatory bowel disease.
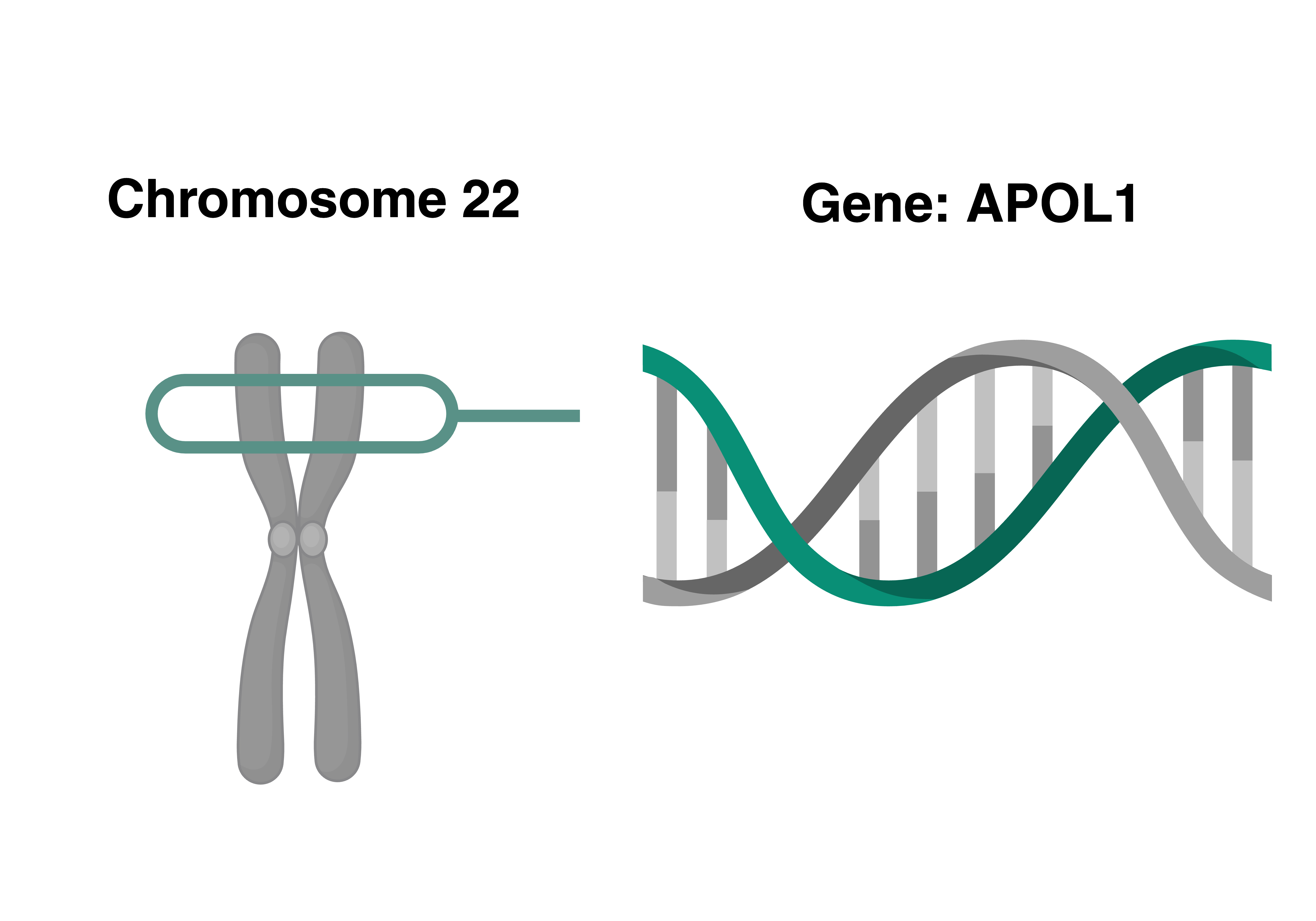
Genetics shape our physiology in complex ways. We use multi-omics data (e.g., DNA methylation, proteomics, metabolomics) to try to understand mechanisms linking genetic risk factors to disease. For example, the variants in the APOL1 gene significantly increase risk for kidney disease, but the pathway between the genetics and disease is unknown. Thus, we have examined associations between APOL1 genetic risk and DNA methylation, metabolomics, and proteomics to try to provide greater insight into the underlying mechanisms.

Human populations are complex with substantial diversity across the globe. The effects of molecular risk factors vary across spectrums of ages, dietary and lifestyle factors, and genetic background. We leverage advanced statistical methods to understand and quantify these differences, e.g., examining interactions between genetic and non-genetic risk factors, and how microbiome profiles and microbiome-disease relationships differ with age, health status, and race/ethnicity.
